Classification: Logistic Regression and Multinomial Extension
Jin Zhu, Liyuan Hu
6/12/2021
Source:vignettes/v03-classification.Rmd
v03-classification.RmdTitanic Dataset and Classification
Consider the Titanic dataset obtained from the Kaggle competition: https://www.kaggle.com/c/titanic/data. The dataset consists of data about 889 passengers, and the goal of the competition is to predict the survival (yes/no) based on features including the class of service, the sex, the age etc.
## [1] 891 12
head(dat)## PassengerId Survived Pclass
## 1 1 0 3
## 2 2 1 1
## 3 3 1 3
## 4 4 1 1
## 5 5 0 3
## 6 6 0 3
## Name Sex Age SibSp Parch
## 1 Braund, Mr. Owen Harris male 22 1 0
## 2 Cumings, Mrs. John Bradley (Florence Briggs Thayer) female 38 1 0
## 3 Heikkinen, Miss. Laina female 26 0 0
## 4 Futrelle, Mrs. Jacques Heath (Lily May Peel) female 35 1 0
## 5 Allen, Mr. William Henry male 35 0 0
## 6 Moran, Mr. James male NA 0 0
## Ticket Fare Cabin Embarked
## 1 A/5 21171 7.2500 <NA> S
## 2 PC 17599 71.2833 C85 C
## 3 STON/O2. 3101282 7.9250 <NA> S
## 4 113803 53.1000 C123 S
## 5 373450 8.0500 <NA> S
## 6 330877 8.4583 <NA> QLogistic regression is one of powerful tool to tackle this problem. In statistics, the logistic regression is used to model the probability of a certain class or event existing such as alive/dead, pass/fail, win/lose, or healthy/sick. Logistic regression function is an -shaped curve modeling the posterior probability via a linear combination of the features. The curve is defined as where and are predictors, and are coefficients to be learned from data. The logistic regression model has this form: The quantity is called the logarithm of the odd, also called log-odd or logit. The best subset selection for logistic regression aim to balance model accuracy and model complexity, where the former is achieves by maximizing the log-likelihood function and the latter is characterized by a constraint: and can be determined in a data driven way.
Best Subset Selection for Logistic Regression
The abess() function in the abess package
allows users to perform best subset selection in a highly efficient way.
Users can call the abess() function using formula just like
what users do with lm(). Or users can specify the design
matrix x and the response y. As an example,
the Titanic dataset is used to demonstrated the usage of
abess package.
Data preprocessing
A glance at the dataset finds there is any missing data. The
na.omit() function allows us to delete the rows that
contain any missing data. After that, we get a total of 714 samples
left.
## [1] 712 8Then we change the factors into dummy variables with the
model.matrix() function. Note that the abess
function will automatically include the intercept, and thus, we exclude
the first column of dat object.
dat <- model.matrix(~., dat)[, -1]
dat <- as.data.frame(dat)We split the dataset into a training set and a test set. The model is going to be built on the training set and later We will test the model performance on the test set.
train_index <- 1:round((712*2)/3)
train <- dat[train_index, ]
test <- dat[-train_index, ]Analyze Titanic dataset with abess package
We use abess package to perform best subset selection
for the preprocessed Titanic dataset by setting
family = "binomial". The cross validation technique is
employed to tune the support size by setting
tune.type = "cv".
library(abess)
abess_fit <- abess(x = train[, -1], y = train$Survived,
family = "binomial", tune.type = "cv")After getting the estimator, we can further do more exploring work.
The output of abess() function contains the best model for
all the candidate support sizes in the support.size. Users
can use some generic functions to quickly draw some information of those
estimators. Typical examples include:
i. print the fitted model:
abess_fit## Call:
## abess.default(x = train[, -1], y = train$Survived, family = "binomial",
## tune.type = "cv")
##
## support.size dev cv
## 1 0 321.9560 64.59866
## 2 1 246.5820 49.99585
## 3 2 229.6078 46.65819
## 4 3 224.0843 47.10831
## 5 4 220.4441 45.75501
## 6 5 220.1834 46.31408
## 7 6 220.0022 46.45407
## 8 7 219.9602 46.39656
## 9 8 219.9210 46.44844- draw the estimated coefficients on all candidate support size by
coef()function:
coef(abess_fit)## 9 x 9 sparse Matrix of class "dgCMatrix"
## 0 1 2 3 4 5
## (intercept) -0.3530971 1.158781 2.9213405 4.23196071 4.76830134 4.85381911
## Pclass . . -0.7952709 -1.01705745 -1.03770046 -1.00849770
## Sexmale . -2.545075 -2.5818944 -2.51198958 -2.60544822 -2.59656529
## Age . . . -0.02923674 -0.03712932 -0.03672237
## SibSp . . . . -0.36190220 -0.35770273
## Parch . . . . . .
## Fare . . . . . .
## EmbarkedQ . . . . . .
## EmbarkedS . . . . . -0.21507393
## 6 7 8
## (intercept) 5.112870189 5.106984422 5.079012311
## Pclass -1.076540803 -1.080872143 -1.085091547
## Sexmale -2.610618146 -2.600190268 -2.601337076
## Age -0.037469295 -0.037121587 -0.037259374
## SibSp -0.339014180 -0.350547546 -0.350406211
## Parch . 0.048198890 0.053009524
## Fare -0.001997874 -0.002237775 -0.002224556
## EmbarkedQ . . 0.210160985
## EmbarkedS -0.245382497 -0.255782092 -0.214986232- get the deviance of the estimated model on all candidate support
sizes via
deviance()function:
deviance(abess_fit)## [1] 321.9560 246.5820 229.6078 224.0843 220.4441 220.1834 220.0022 219.9602
## [9] 219.9210- visualize the change of models with the change of support size via
plot()function:
plot(abess_fit, label=T)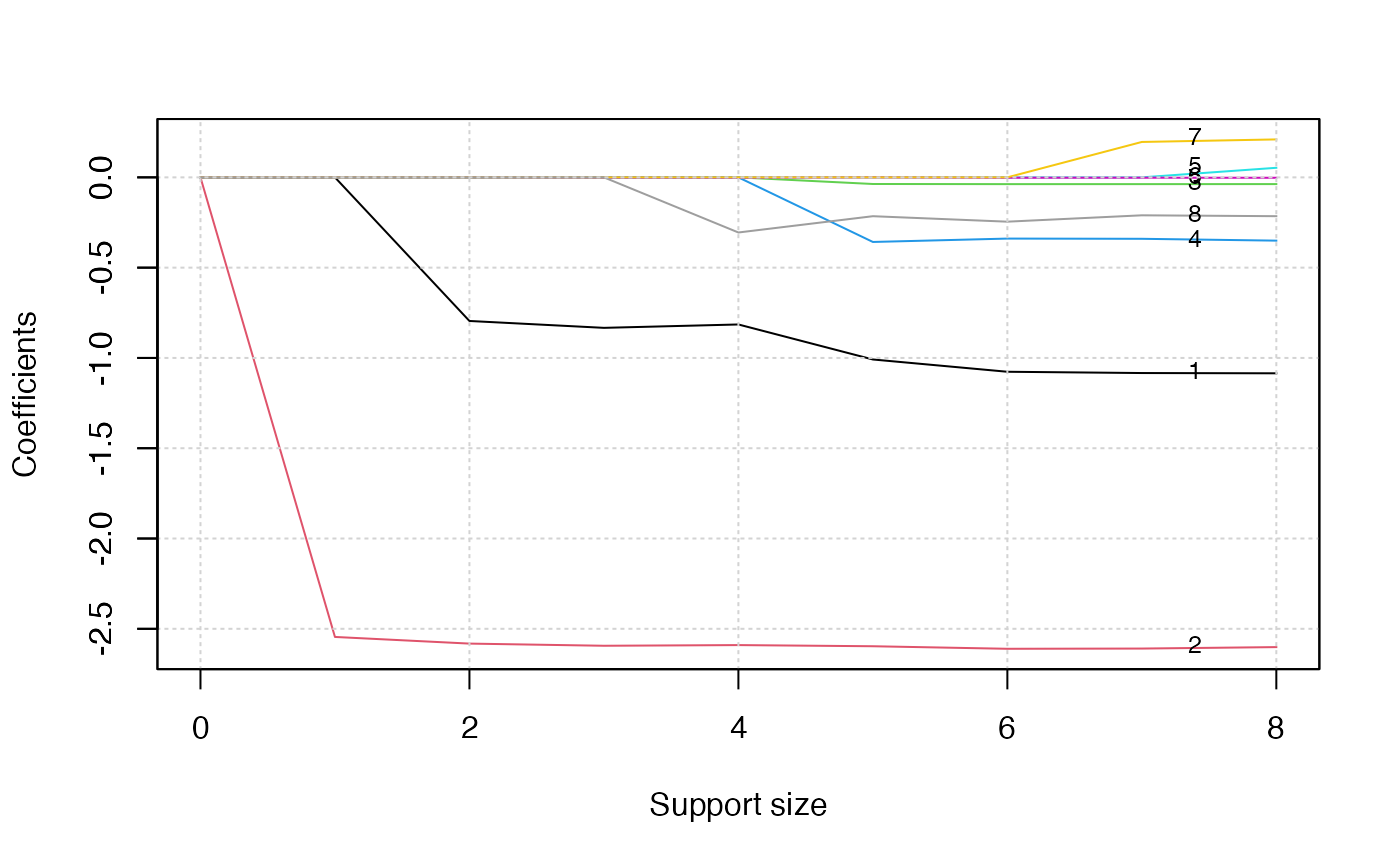
The graph shows that, beginning from the most dense model, the second
variable (Sex) is included in the active set until the
support size reaches 0. We can also generate a graph about the tuning
value.
plot(abess_fit, type = "tune")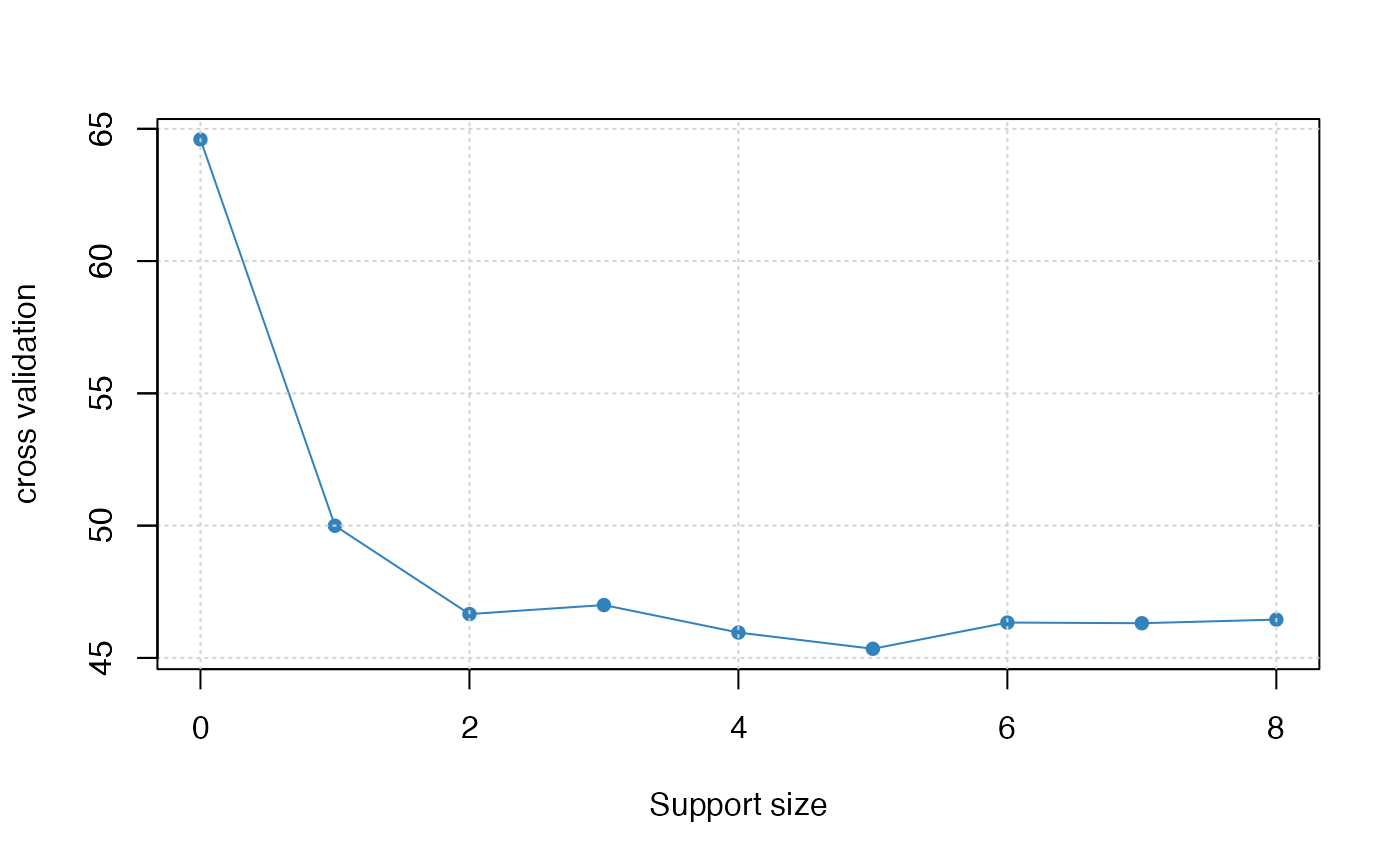
The tuning value reaches the lowest point at 4, which implies the
best model consists of four variables.
Finally, to extract the specified model from the abess
object, we can call the extract() function with a given
support.size. If support.size is not provided,
the model with the best tuning value will be returned. Here we extract
the model with support size equals 6.
## List of 7
## $ beta :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## .. ..@ i : int [1:4] 0 1 2 3
## .. ..@ p : int [1:2] 0 4
## .. ..@ Dim : int [1:2] 8 1
## .. ..@ Dimnames:List of 2
## .. .. ..$ : chr [1:8] "Pclass" "Sexmale" "Age" "SibSp" ...
## .. .. ..$ : chr "4"
## .. ..@ x : num [1:4] -1.0377 -2.6054 -0.0371 -0.3619
## .. ..@ factors : list()
## $ intercept : num 4.77
## $ support.size: num 4
## $ support.vars: chr [1:4] "Pclass" "Sexmale" "Age" "SibSp"
## $ support.beta: num [1:4] -1.0377 -2.6054 -0.0371 -0.3619
## $ dev : num 220
## $ tune.value : num 45.8The return is a list containing the basic information of the estimated model.
Make a Prediction
Prediction is allowed for all the estimated models. Just call
predict.abess() function with the support.size
set to the size of model users are interested in. If
support.size is not provided, prediction will be made based
on the best tuning value. The predict.abess() can provide
both link, standing for the linear predictors, and the
response, standing for the fitted probability. Here We will
predict the probability of survival on the test.csv
data.
fitted.results <- predict(abess_fit, newx = test, type = 'response')If we chose 0.5 as the cut point, i.e, we predict the person survives the sinking of the Titanic if the fitted probability is greater than 0.5, the accuracy will be 0.80.
fitted.results <- ifelse(fitted.results > 0.5, 1, 0)
misClasificError <- mean(fitted.results != test$Survived)
print(paste('Accuracy',1-misClasificError))## [1] "Accuracy 0.79746835443038"We can also generate an ROC and calculate the AUC value. On this dataset, the AUC is 0.87, which is quite close to 1.
library(ROCR)
fitted.results <- predict(abess_fit, newx = test, type = 'response')
pr <- prediction(fitted.results, test$Survived)
prf <- performance(pr, measure = "tpr", x.measure = "fpr")
plot(prf)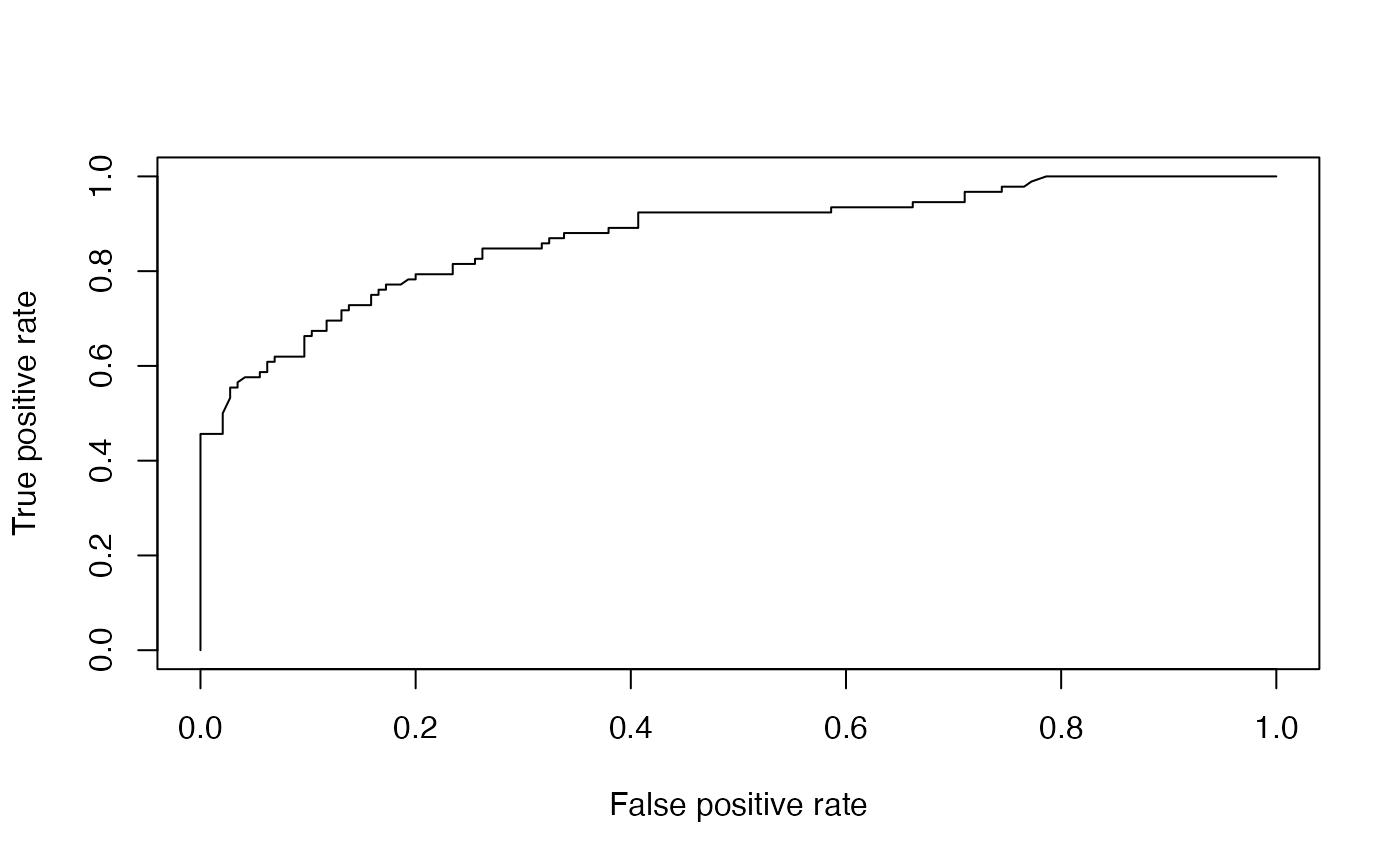
auc <- performance(pr, measure = "auc")
auc <- auc@y.values[[1]]
auc## [1] 0.8739505Extension: Multi-class Classification
Best subset selection for multinomial logistic regression
When the number of classes is more than 2, we call it multi-class classification task. Logistic regression can be extended to model several classes of events such as determining whether an image contains a cat, dog, lion, etc. Each object detected in the image would be assigned a probability between 0 and 1, with a sum of one. The extended model is multinomial logistic regression.
To arrive at the multinomial logistic model, one can imagine, for possible classes, running independent logistic regression models, in which one class is chosen as a ``pivot’’ and then the other classes are separately regressed against the pivot outcome. This would proceed as follows, if class K (the last outcome) is chosen as the pivot: Then, the probability to choose the -th class can be easily derived to be: and subsequently, we would predict the -th class if the . Notice that, for possible classes case, there are unknown parameters: to be estimated. Because the number of parameters increase as , it is even more urge to constrain the model complexity. And the best subset selection for multinomial logistic regression aims to maximize the log-likelihood function and control the model complexity by restricting with where $\| B \|_{0, 2} = \sum_{i = 1}^{p} I(B_{i\cdot} = {\bf 0})$, is the -th row of coefficient matrix and ${\bf 0} \in R^{K - 1}$ is an all zero vector. In other words, each row of would be either all zero or all non-zero.
Multinomial logistic regression with abess Package
We shall conduct Multinomial logistic regression on an artificial
dataset for demonstration. The generate.data() function
provides a simple way to generate suitable for this task. The assumption
behind is the response vector following a multinomial distribution. The
artifical dataset contain 100 observations and 20 predictors but only
five predictors have influence on the three possible classes.
library(abess)
n <- 100
p <- 20
support.size <- 5
dataset <- generate.data(n, p, support.size, family = "multinomial", class.num = 3)
head(dataset$y)## [1] 1 2 1 1 0 0
dataset$beta## [,1] [,2] [,3]
## [1,] 0.000000 0.000000 0.000000
## [2,] 0.000000 0.000000 0.000000
## [3,] 0.000000 0.000000 0.000000
## [4,] 0.000000 0.000000 0.000000
## [5,] 0.000000 0.000000 0.000000
## [6,] 16.244248 6.605285 4.958967
## [7,] 0.000000 0.000000 0.000000
## [8,] 0.000000 0.000000 0.000000
## [9,] 0.000000 0.000000 0.000000
## [10,] 0.000000 0.000000 0.000000
## [11,] 3.462001 6.774016 20.454220
## [12,] 0.000000 0.000000 0.000000
## [13,] 18.987979 13.133145 17.211335
## [14,] 8.034160 1.117907 2.904334
## [15,] 0.000000 0.000000 0.000000
## [16,] 0.000000 0.000000 0.000000
## [17,] 0.000000 0.000000 0.000000
## [18,] 23.307996 17.576156 11.922882
## [19,] 0.000000 0.000000 0.000000
## [20,] 0.000000 0.000000 0.000000To carry out best subset selection for multinomial logistic
regression, users can call the abess() function with
family specified to multinomial. Here is an
example.
abess_fit <- abess(dataset[["x"]], dataset[["y"]],
family = "multinomial", tune.type = "cv")
extract(abess_fit)[["support.vars"]]## [1] "x6" "x11" "x13" "x14" "x18"Notice that the abess() correctly identifies the support
set of the ground truth coefficient matrix.