Tips for faster computation
Jin Zhu
6/13/2021
Source:vignettes/v09-fasterSetting.Rmd
v09-fasterSetting.RmdIntroduction
The generic splicing technique certifiably guarantees the best subset can be selected in a polynomial time. In practice, the computational efficiency can be improved to handle large scale datasets. The tips for computational improvement include:
- exploit sparse structure of input matrix;
- use golden-section to search best support size;
- early-stop scheme;
- sure independence screening;
- warm-start initialization;
- parallel computing when performing cross validation;
- covariance update for
family = "gaussian"orfamily = "mgaussian";
- approximate Newton iteration for
family = "binomial",family = "poisson",family = "cox".
This vignette illustrate the first three tips. For the other tips, they have been efficiently implemented and set as the default in abess package.
Sparse matrix
We sometimes meet with problems where the input matrix is extremely sparse, i.e., many entries in have zero values. A notable example comes from document classification: aiming to assign classes to a document, making it easier to manage for publishers and news sites. The input variables for characterizing documents are generated from a so called ``bag-of-words’’ model. In this model, each variable is scored for the presence of each of the words in the entire dictionary under consideration. Since most words are absent, the input variables for each document is mostly zero, and so the entire matrix is mostly zero. Such sparse matrices can be efficiently stored in R with a sparse column format via the Matrix package. And the sparse matrix can be directly used by our abess package for boosting the computational efficient.
ABESS algorithm is ideally set up to exploit such sparsity. The inner-product operations when computing forward sacrifice can exploit the sparsity, by summing over only the non-zero entries. For computing backward sacrifice, the sparsity also facilitate solving the convex optimization under a given support set. The following example demonstrates the efficiency gain from the sparse matrix. We first generate a input matrix whose 90% entries are 0.
## Loading required package: Matrix
num <- 1000
p <- 100
sparse_ratio <- 0.9
x <- matrix(rnorm(num * p), nrow = num)
zero_entry <- matrix(rbinom(num * p, size = 1, prob = sparse_ratio), nrow = num)
x[zero_entry == 1] <- 0
y <- x %*% matrix(c(rep(3, 5), rep(0, p - 5)))
x <- Matrix(x)
head(x[, 1:10])## 6 x 10 sparse Matrix of class "dgCMatrix"
##
## [1,] . . . 1.120616 . . . . . .
## [2,] . . . . . . . . . .
## [3,] . . . . . . . . . .
## [4,] . . . . . . . . . .
## [5,] . . . . . . . . 0.09618829 .
## [6,] . . . . . . 1.12201 . . .Then, we apply ABESS algorithm on Matrix x and record
the runtime in t1:
##
## Thank you for using abess! To acknowledge our work, please cite the package:##
## Zhu J, Wang X, Hu L, Huang J, Jiang K, Zhang Y, Lin S, Zhu J (2022). 'abess: A Fast Best Subset Selection Library in Python and R.' Journal of Machine Learning Research, 23(202), 1-7. https://www.jmlr.org/papers/v23/21-1060.html.
t1 <- system.time(abess_fit <- abess(x, y))We compare the runtime when the input matrix is dense matrix:
## [,1] [,2] [,3] [,4] [,5] [,6]
## [1,] 0 0 0 1.120616 0 0
## [2,] 0 0 0 0.000000 0 0
## [3,] 0 0 0 0.000000 0 0
## [4,] 0 0 0 0.000000 0 0
## [5,] 0 0 0 0.000000 0 0
## [6,] 0 0 0 0.000000 0 0
t2 <- system.time(abess_fit <- abess(x, y))
rbind(t1, t2)[, 1:3]## user.self sys.self elapsed
## t1 0.190 0.004 0.198
## t2 0.196 0.022 0.230From the comparison, we see that the time required by sparse matrix
is visibly smaller, and thus, we suggest to assign a sparse matrix to
abess when the input matrix have a lot of zero entries.
Golden-section searching
The following is a typical ``model size v.s. BGIC’’ plot.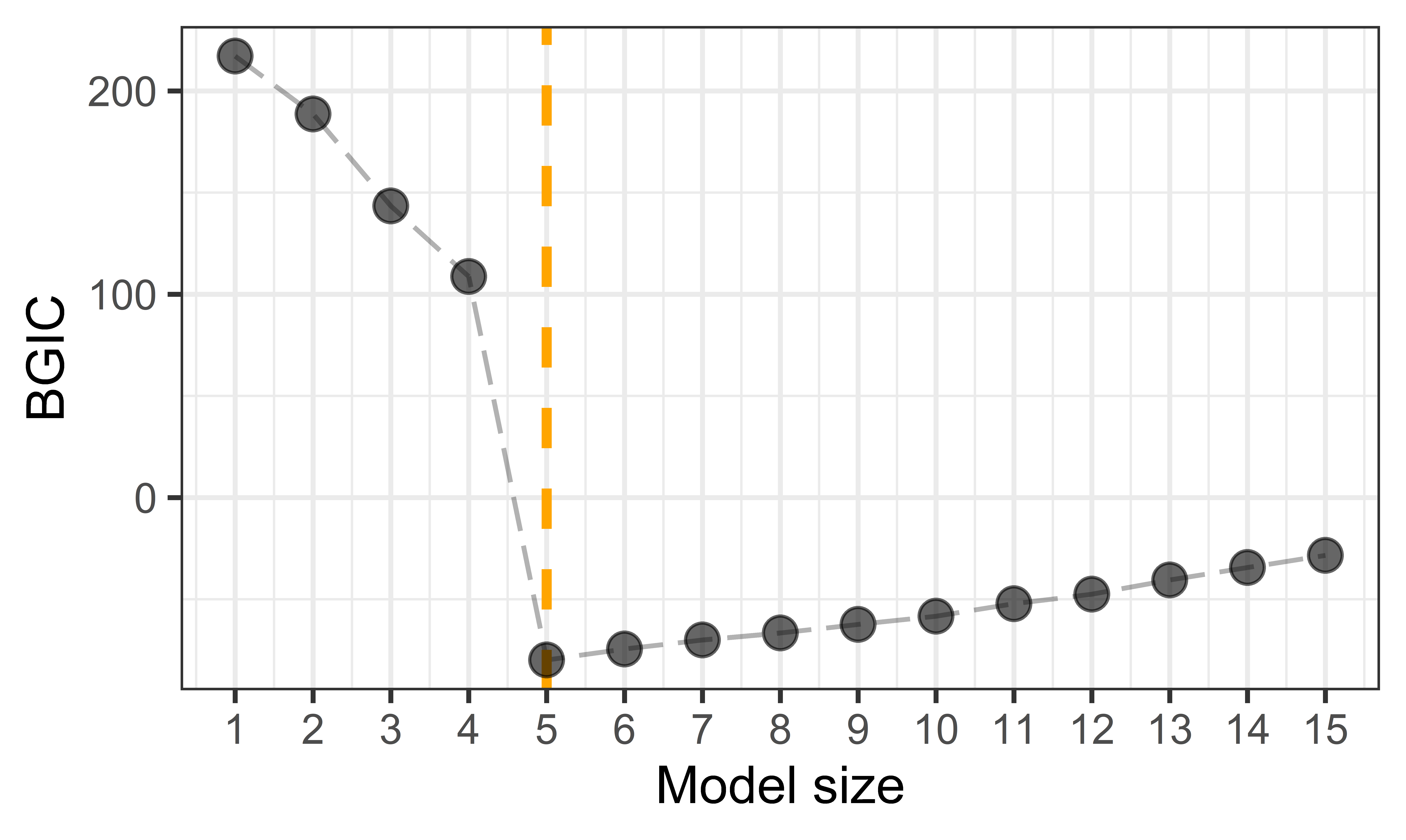
The -axis is model size, and the -axis is BGIC’s value recorded in group splicing algorithm for linear model. The entries of design matrix are i.i.d. sampled from , and the matrix shape is . The error term are i.i.d. . Take the two adjacent variables as one group, and set the true coefficients . The orange vertical dash line indicates the true group subset size.
From this Figure, we see that the BGIC decreases from to , but it increases as larger than . In other words, the BGIC path of SGSplicing algorithm is a strictly unimodal function achieving minimum at the true group subset size . Motivated by this observation, we suggest to recruit a heuristic search based on the golden-section search technique, an efficient method for finding the extremum of a unimodal function, to determine support size that minimizing BGIC. Compared with searching the optimal support size one by one from a candidate set with complexity, golden-section reduce the time complexity to , giving a significant computational improvement.
The code below exhibits how to employ the golden search technique with abess package:
synthetic_data <- generate.data(n = 500, p = 100,
beta = c(3, 1.5, 0, 0, 2, rep(0, 95)))
dat <- cbind.data.frame("y" = synthetic_data[["y"]],
synthetic_data[["x"]])
t1 <- system.time(abess_fit <- abess(y ~ ., data = dat, tune.path = "gsection"))
str(extract(abess_fit))## List of 7
## $ beta :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## .. ..@ i : int [1:3] 0 1 4
## .. ..@ p : int [1:2] 0 3
## .. ..@ Dim : int [1:2] 100 1
## .. ..@ Dimnames:List of 2
## .. .. ..$ : chr [1:100] "x1" "x2" "x3" "x4" ...
## .. .. ..$ : chr "3"
## .. ..@ x : num [1:3] 3.04 1.49 1.91
## .. ..@ factors : list()
## $ intercept : num -0.0679
## $ support.size: int 3
## $ support.vars: chr [1:3] "x1" "x2" "x5"
## $ support.beta: num [1:3] 3.04 1.49 1.91
## $ dev : num 0.745
## $ tune.value : num -122The output of golden-section strategy suggests the optimal model size is accurately detected. Compare to the sequential searching, the golden section reduce the runtime because it skip some support sizes which are likely to be a non-optimal one:
t2 <- system.time(abess_fit <- abess(y ~ ., data = dat))
rbind(t1, t2)[, 1:3]## user.self sys.self elapsed
## t1 0.020 0.001 0.024
## t2 0.038 0.002 0.046Early stop
In machine learning, early stopping is a helpful strategy not only avoid overfitting but also reducing training time. For the early-stopping implementation in abess, validation is used to detect when overfitting starts during performing adaptive best subset selection; training is then stopped even though the best subset under certain larger support size haven’t found. We give an example to demonstrate the helpfulness of early stopping in decreasing runtimes. (Do not finish, the early stopping do not available in cpp)
t1 <- system.time(abess_fit <- abess(y ~ ., data = dat, early.stop = TRUE))
abess_fit## Call:
## abess.formula(formula = y ~ ., data = dat, early.stop = TRUE)
##
## support.size dev GIC
## 1 0 8.6400493 1078.204142
## 2 1 3.8958639 688.370921
## 3 2 1.9816733 358.797191
## 4 3 0.7451024 -121.877226
## 5 4 0.7337883 -121.114561
## 6 5 0.7229702 -120.127663
## 7 6 0.7163121 -116.340488
## 8 7 0.7091789 -112.931327
## 9 8 0.7024621 -109.276323
## 10 9 0.6976621 -104.291399
## 11 10 0.6932543 -99.047213
## 12 11 0.6892982 -93.495441
## 13 12 0.6846565 -88.460620
## 14 13 0.6810295 -82.703248
## 15 14 0.6772793 -77.051013
## 16 15 0.6741793 -70.931592
## 17 16 0.6710871 -64.817014
## 18 17 0.6682440 -58.526563
## 19 18 0.6653608 -52.275367
## 20 19 0.6628664 -45.740178
## 21 20 0.6603517 -39.227384
## 22 21 0.6578908 -32.681006
## 23 22 0.6555246 -26.069380
## 24 23 0.6535097 -19.195392
## 25 24 0.6515939 -12.250127
## 26 25 0.6494848 -5.457983
## 27 26 0.6476394 1.532509
## 28 27 0.6457393 8.476659
## 29 28 0.6438120 15.395256
## 30 29 0.6418701 22.298056
## 31 30 0.6397735 29.075429
## 32 31 0.6380951 36.175194
## 33 32 0.6363094 43.187138
## 34 33 0.6345171 50.189989
## 35 34 0.6327394 57.200441
## 36 35 0.6310138 64.248152
## 37 36 0.6294229 71.399182
## 38 37 0.6278000 78.521521
## 39 38 0.6264222 85.836180
## 40 39 0.6242619 92.522044
## 41 40 0.6223307 99.386095
## 42 41 0.6210093 106.736478
## 43 42 0.6197906 114.167492
## 44 43 0.6185396 121.570460
## 45 44 0.6172515 128.941329
## 46 45 0.6159546 136.302901
## 47 46 0.6147094 143.704246
## 48 47 0.6137414 151.329447
## 49 48 0.6127953 158.971283
## 50 49 0.6118767 166.634416
## 51 50 0.6110400 174.363418
## 52 51 0.6101414 182.040795
## 53 52 0.6094084 189.853000
## 54 53 0.6086294 197.626589
## 55 54 0.6077610 205.325872
## 56 55 0.6070552 213.158128
## 57 56 0.6062506 220.908182
## 58 57 0.6054073 228.625357
## 59 58 0.6046684 236.427918
## 60 59 0.6039253 244.226313We can see that ABESS algorithm stop when support size is 4. This is because the GIC value (can be considered as an assessment in validation set) do not increase when support size reach to 3, and thus, the program early terminate. This result is match to our simulation setting. Compare with the ABESS without early-stopping:
t2 <- system.time(abess_fit <- abess(y ~ ., data = dat))
rbind(t1, t2)[, 1:3]## user.self sys.self elapsed
## t1 0.037 0.002 0.042
## t2 0.029 0.002 0.031we can conclude that early-stopping brings fast computation and might maintain statistical guarantee.