Advanced Features
Jin Zhu, Yanhang Zhang
1/21/2022
Source:vignettes/v07-advancedFeatures.Rmd
v07-advancedFeatures.RmdIntroduction
When analyzing the real world datasets, we may have the following
targets:
1. certain variables must be selected when some prior information is
given;
2. selecting the weak signal variables when the prediction performance
is main interest;
3. identifying predictors when group structure are provided;
4. pre-excluding a part of predictors when datasets have ultra
high-dimensional predictors.
In the following content, we will illustrate the statistic methods to
reach these targets in a one-by-one manner, and give quick examples to
show how to perform the statistic methods with the abess
package. Actually, in the abess package, there targets can
be properly handled by simply change the default arguments in the
abess() function.
Nuisance Regression
Nuisance regression refers to best subset selection with some prior information that some variables are required to stay in the active set. For example, if we are interested in a certain gene and want to find out what other genes are associated with the response when this particular gene shows effect.
Carrying Out the Nuisance Regression with abess
In the abess() function, the argument
always.include is designed to realize this goal. user can
pass a vector containing the indexes of the target variables to
always.include. Here is an example.
##
## Thank you for using abess! To acknowledge our work, please cite the package:##
## Zhu J, Wang X, Hu L, Huang J, Jiang K, Zhang Y, Lin S, Zhu J (2022). 'abess: A Fast Best Subset Selection Library in Python and R.' Journal of Machine Learning Research, 23(202), 1-7. https://www.jmlr.org/papers/v23/21-1060.html.
n <- 100
p <- 20
support.size <- 3
dataset <- generate.data(n, p, support.size)
dataset$beta## [1] 0.00000 0.00000 0.00000 0.00000 0.00000 84.67713 0.00000 0.00000
## [9] 0.00000 0.00000 0.00000 0.00000 0.00000 85.82187 0.00000 0.00000
## [17] 0.00000 47.40922 0.00000 0.00000
abess_fit <- abess(dataset[["x"]], dataset[["y"]], always.include = 6)
coef(abess_fit, support.size = abess_fit$support.size[which.min(abess_fit$tune.value)])## 21 x 1 sparse Matrix of class "dgCMatrix"
## 4
## (intercept) -4.767241
## x1 .
## x2 .
## x3 .
## x4 .
## x5 .
## x6 82.424118
## x7 .
## x8 .
## x9 .
## x10 .
## x11 .
## x12 .
## x13 .
## x14 89.031689
## x15 .
## x16 -12.622583
## x17 .
## x18 43.343247
## x19 .
## x20 .Regularized Adaptive Best Subset Selection
In some cases, especially under low signal-to-noise ratio (SNR)
setting or predictors are highly correlated, the vanilla type of
constrained model may not be satisfying and a more sophisticated
trade-off between bias and variance is needed. Under this concern, the
abess package provides option of best subset selection with
norm regularization called the regularized best subset selection (BESS).
The model has this following form:
Carrying out the Regularized BESS
To implement the regularized BESS, user need to specify a value to
the lambda in the abess() function. This
lambda value corresponds to the penalization parameter in
the model above. Here we give an example.
library(abess)
n <- 100
p <- 30
snr <- 0.05
dataset <- generate.data(n, p, snr = snr, seed = 1, beta = rep(c(1, rep(0 ,5)), each = 5), rho = 0.8, cortype = 3)
data.test <- generate.data(n, p, snr = snr, beta = dataset$beta, seed = 100)
abess_fit <- abess(dataset[["x"]], dataset[["y"]], lambda = 0.7)Let’s test the regularized best subset selection against the no-regularized one over 100 replications in terms of prediction performance.
M <- 100
err.l0 <- rep(0, M)
err.l0l2 <- rep(0, M)
for(i in 1:M){
dataset <- generate.data(n, p, snr = snr, seed = i, beta = rep(c(1, rep(0 ,5)), each = 5), rho = 0.8, cortype = 3)
data.test <- generate.data(n, p, snr = snr, beta = dataset$beta, seed = i+100)
abess_fit <- abess(dataset[["x"]], dataset[["y"]], lambda = 0.7)
coef(abess_fit, support.size = abess_fit$support.size[which.min(abess_fit$tune.value)])
pe.l0l2 <- norm(data.test$y - predict(abess_fit, newx = data.test$x),'2')
err.l0[i] <- pe.l0l2
abess_l0 <- abess(dataset[["x"]], dataset[["y"]])
coef(abess_l0, support.size = abess_l0$support.size[which.min(abess_l0$tune.value)])
pe.l0 <- norm(data.test$y -predict(abess_l0, newx = data.test$x), '2')
err.l0l2[i] <- pe.l0
}
mean(err.l0)## [1] 132.5536
mean(err.l0l2)## [1] 151.622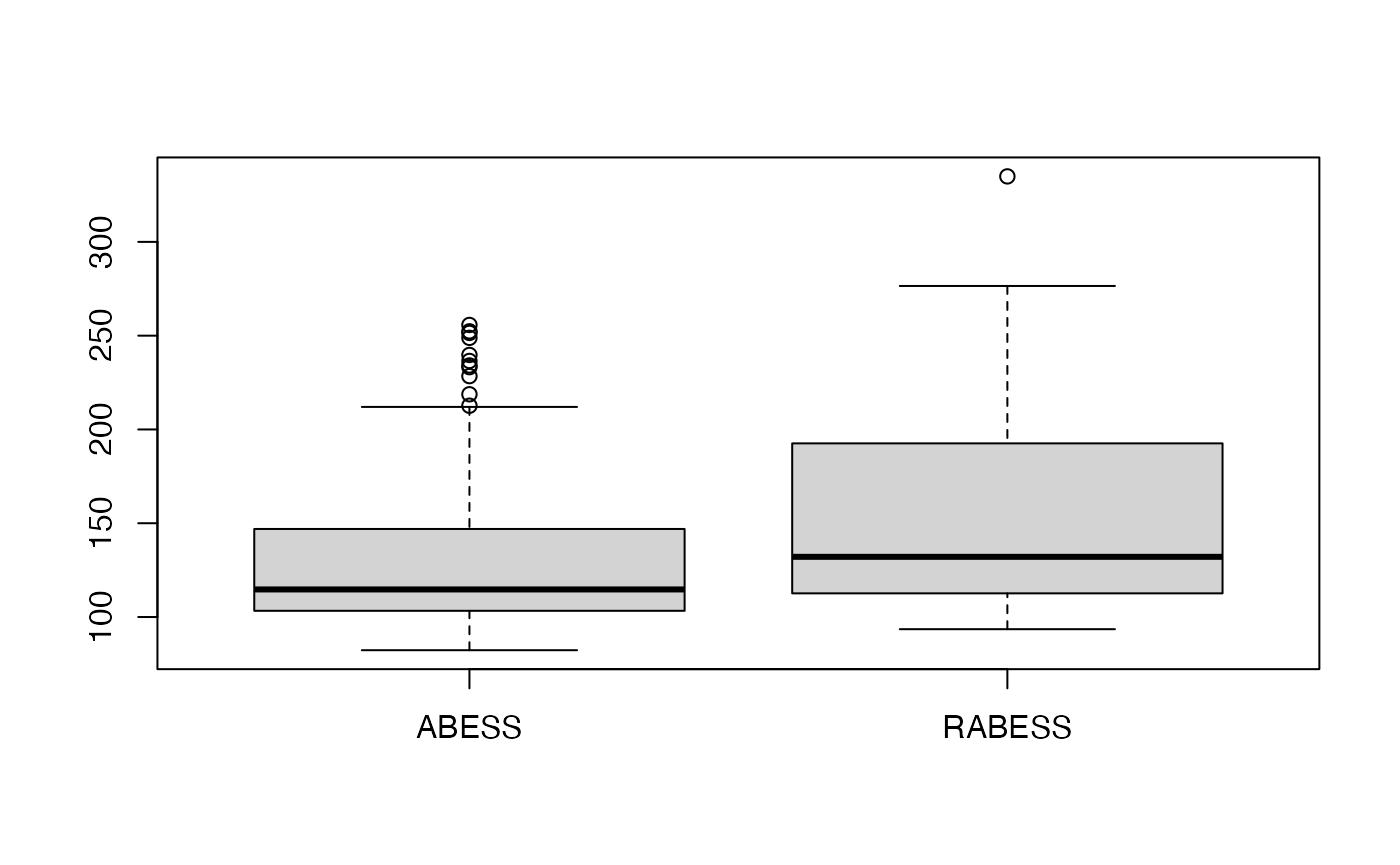
We see that the regularized best subset select indeed reduces the prediction error.
Best group subset selection
Group linear model is a linear model which the predictors are separated into pre-determined non-overlapping groups,
where are the group indices of the predictors.
Best group subset selection (BGSS) aims to choose a small part of groups to achieve the best interpretability on the response variable. BGSS is practically useful for the analysis of variables with certain group structures.
The BGSS can be achieved by solving:
where the
(-pseudo) norm is defined as
and model size
is a positive integer to be determined from data. Regardless of the
NP-hard of this problem, Zhang et al develop a certifiably polynomial
algorithm with a high probability to solve it. This algorithm is
integrated in the abess package, and user can handily
select best group subset by assigning a proper value to the
group.index arguments in abess() function.
Quick example
We use a synthetic dataset to demonstrate its usage. The dataset consists of 100 observations. Each observation has 20 predictors but only the first three predictors among them have a impact on the response.
set.seed(1234)
n <- 100
p <- 20
support.size <- 3
dataset <- generate.data(n, p, beta = c(10, 5, 5, rep(0, 17)))Supposing we have some prior information that the second and third
variables belong to the same group, then we can set the
group_index as:
Then, pass this group_index to the abess()
function:
abess_fit <- abess(dataset[["x"]], dataset[["y"]],
group.index = group_index)
str(extract(abess_fit))## List of 7
## $ beta :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## .. ..@ i : int [1:3] 0 1 2
## .. ..@ p : int [1:2] 0 3
## .. ..@ Dim : int [1:2] 20 1
## .. ..@ Dimnames:List of 2
## .. .. ..$ : chr [1:20] "x1" "x2" "x3" "x4" ...
## .. .. ..$ : chr "2"
## .. ..@ x : num [1:3] 10.2 4.92 4.88
## .. ..@ factors : list()
## $ intercept : num 0.0859
## $ support.size: int 2
## $ support.vars: chr [1:3] "x1" "x2" "x3"
## $ support.beta: num [1:3] 10.2 4.92 4.88
## $ dev : num 8.3
## $ tune.value : num 225The fitted result suggest that only two groups are selected (since
support.size is 2) and the selected variables are matched
with the ground truth setting.
Feature screening
In the case of ultra high-dimensional data, it is more effective to
do feature screening first before carrying out feature selection. The
abess package allowed users to perform the sure independent
screening to pre-exclude some features according to the marginal
likelihood.
Integrate SIS in abess package
Ultra-high dimensional predictors increase computational cost but
reduce estimation accuracy for any statistical procedure. To reduce
dimensionality from high to a relatively acceptable level, a fairly
general asymptotic framework, named feature screening (sure independence
screening) is proposed to tackle even exponentially growing dimension.
The feature screening can theoretically maintain all effective
predictors with a high probability, which is called the sure screening
property. In the abess() function, to carrying out the
Integrate SIS, user need to passed an integer smaller than the number of
the predictors to the screening.num. Then the
abess() function will first calculate the marginal
likelihood of each predictor and reserve those predictors with the
screening.num largest marginal likelihood. Then, the ABESS
algorithm is conducted only on this screened subset. Here is an
example.
library(abess)
n <- 100
p <- 1000
support.size <- 3
dataset <- generate.data(n, p, support.size)
abess_fit <- abess(dataset[["x"]], dataset[["y"]],
screening.num = 100)